Module 2: Bonus Exercise Results
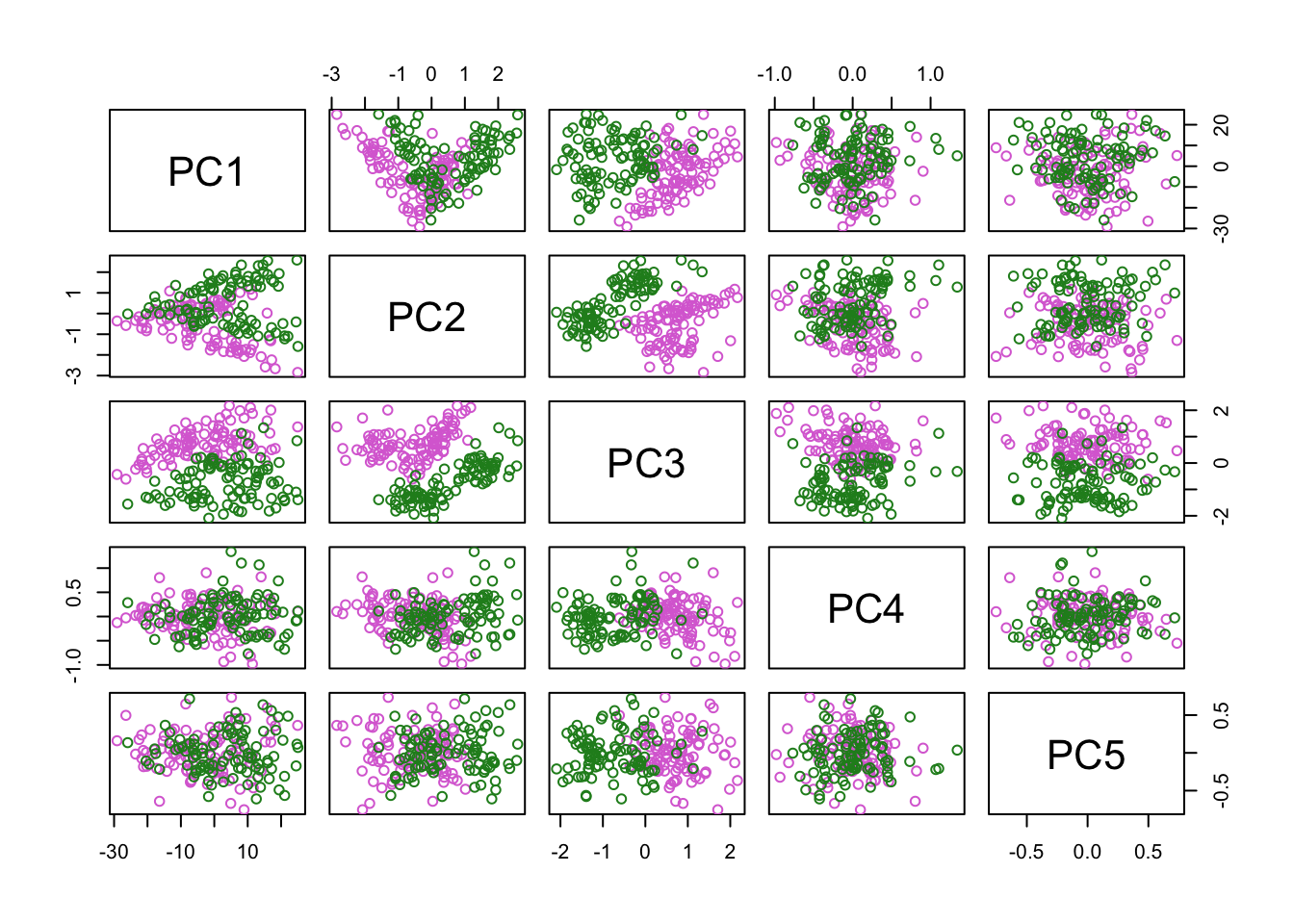
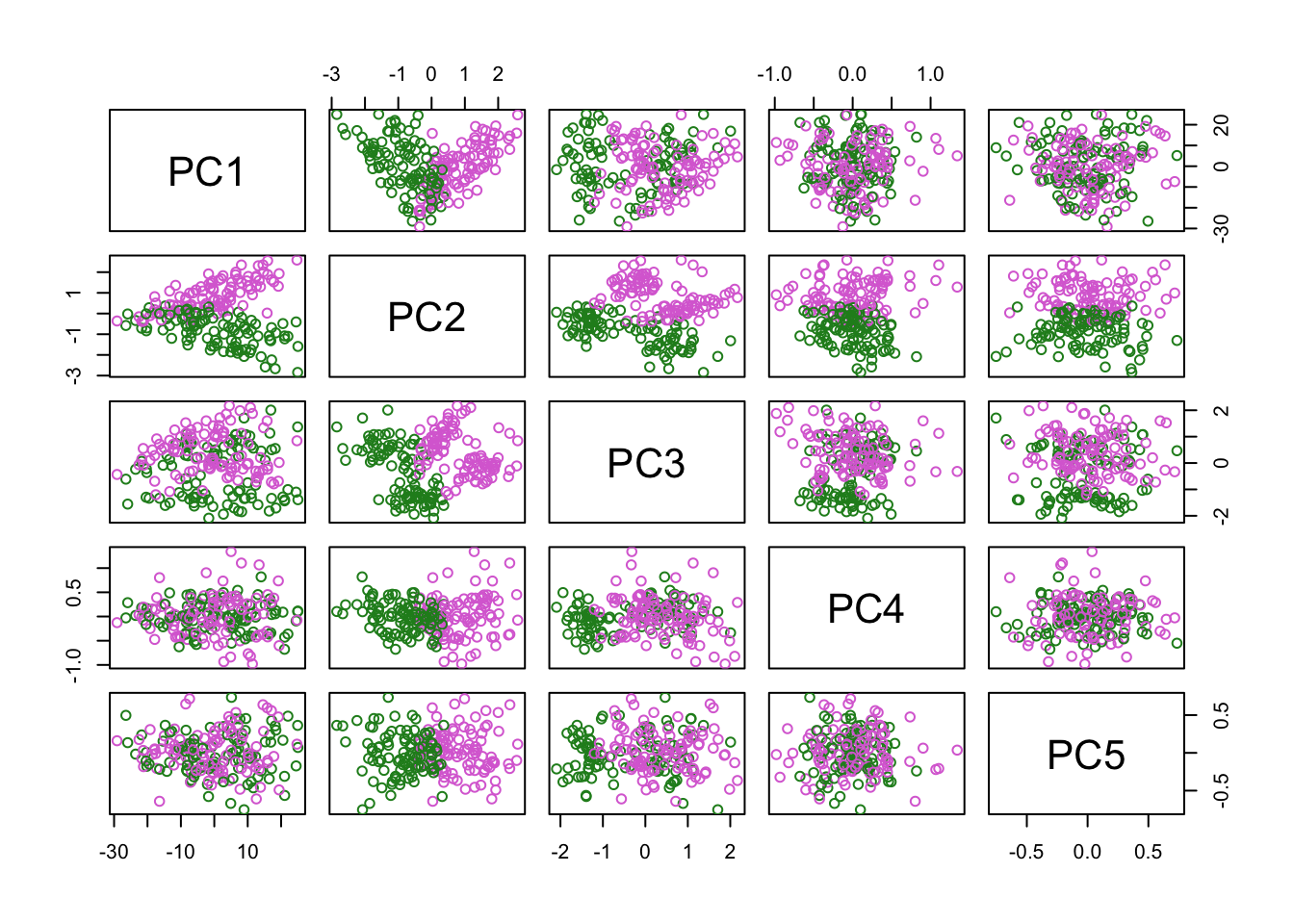
PCA
Plot PC projections (embeddings).
tSNE:
## sigma summary: Min. : 0.295392306171995 |1st Qu. : 0.424864940106807 |Median : 0.475900590252246 |Mean : 0.477263744443299 |3rd Qu. : 0.522810659014478 |Max. : 0.672971536327323 |## Epoch: Iteration #100 error is: 14.7258175926178## Epoch: Iteration #200 error is: 0.445766486822539## Epoch: Iteration #300 error is: 0.411654267486382## Epoch: Iteration #400 error is: 0.40309763203543## Epoch: Iteration #500 error is: 0.399613102555782## Epoch: Iteration #600 error is: 0.397408371315958## Epoch: Iteration #700 error is: 0.395992460654595## Epoch: Iteration #800 error is: 0.394855618443548## Epoch: Iteration #900 error is: 0.393961257756425## Epoch: Iteration #1000 error is: 0.393215111303993## sigma summary: Min. : 0.42069998064187 |1st Qu. : 0.505494820242659 |Median : 0.550282641638609 |Mean : 0.553782538032253 |3rd Qu. : 0.597446288884567 |Max. : 0.737568418500652 |## Epoch: Iteration #100 error is: 13.7341255620951## Epoch: Iteration #200 error is: 0.397095487609806## Epoch: Iteration #300 error is: 0.350675482216564## Epoch: Iteration #400 error is: 0.347798500856526## Epoch: Iteration #500 error is: 0.346750213689565## Epoch: Iteration #600 error is: 0.346207721864965## Epoch: Iteration #700 error is: 0.345856707586482## Epoch: Iteration #800 error is: 0.345615067626572## Epoch: Iteration #900 error is: 0.34543303007688## Epoch: Iteration #1000 error is: 0.34528845000959## sigma summary: Min. : 0.539839363698465 |1st Qu. : 0.634067694694373 |Median : 0.675230651916411 |Mean : 0.676426601512199 |3rd Qu. : 0.712708887622463 |Max. : 0.85041386579969 |## Epoch: Iteration #100 error is: 13.6251777853525## Epoch: Iteration #200 error is: 0.338825684375918## Epoch: Iteration #300 error is: 0.311984418755623## Epoch: Iteration #400 error is: 0.311106848807463## Epoch: Iteration #500 error is: 0.311106848487757## Epoch: Iteration #600 error is: 0.311106848487757## Epoch: Iteration #700 error is: 0.311106848487757## Epoch: Iteration #800 error is: 0.311106848487757## Epoch: Iteration #900 error is: 0.311106848487757## Epoch: Iteration #1000 error is: 0.311106848487757## sigma summary: Min. : 0.689338665294285 |1st Qu. : 0.801156853023062 |Median : 0.838030059692607 |Mean : 0.83585263946599 |3rd Qu. : 0.869043547272454 |Max. : 1.00462478171883 |## Epoch: Iteration #100 error is: 10.7616876869579## Epoch: Iteration #200 error is: 0.275011673610231## Epoch: Iteration #300 error is: 0.275010788573918## Epoch: Iteration #400 error is: 0.275010788573788## Epoch: Iteration #500 error is: 0.275010788573788## Epoch: Iteration #600 error is: 0.275010788573788## Epoch: Iteration #700 error is: 0.275010788573788## Epoch: Iteration #800 error is: 0.275010788573788## Epoch: Iteration #900 error is: 0.275010788573788## Epoch: Iteration #1000 error is: 0.275010788573788sex_cols = c(“orchid”,“forestgreen”)[factor(crabs$sex)]
Color-code tSNE plot by species, try various perplexity levels:
species_cols = c("orchid","forestgreen")[factor(crabs$sp)]
par(mfrow=c(2,2))
plot(c_tsne10[,1],
c_tsne10[,2],
main = "Perplexity = 10",
col = species_cols)
plot(c_tsne20[,1],
c_tsne20[,2],
main = "Perplexity = 20",
col = species_cols)
plot(c_tsne50[,1],
c_tsne50[,2],
main = "Perplexity = 50",
col = species_cols)
plot(c_tsne100[,1],
c_tsne100[,2],
main = "Perplexity = 100",
col = species_cols)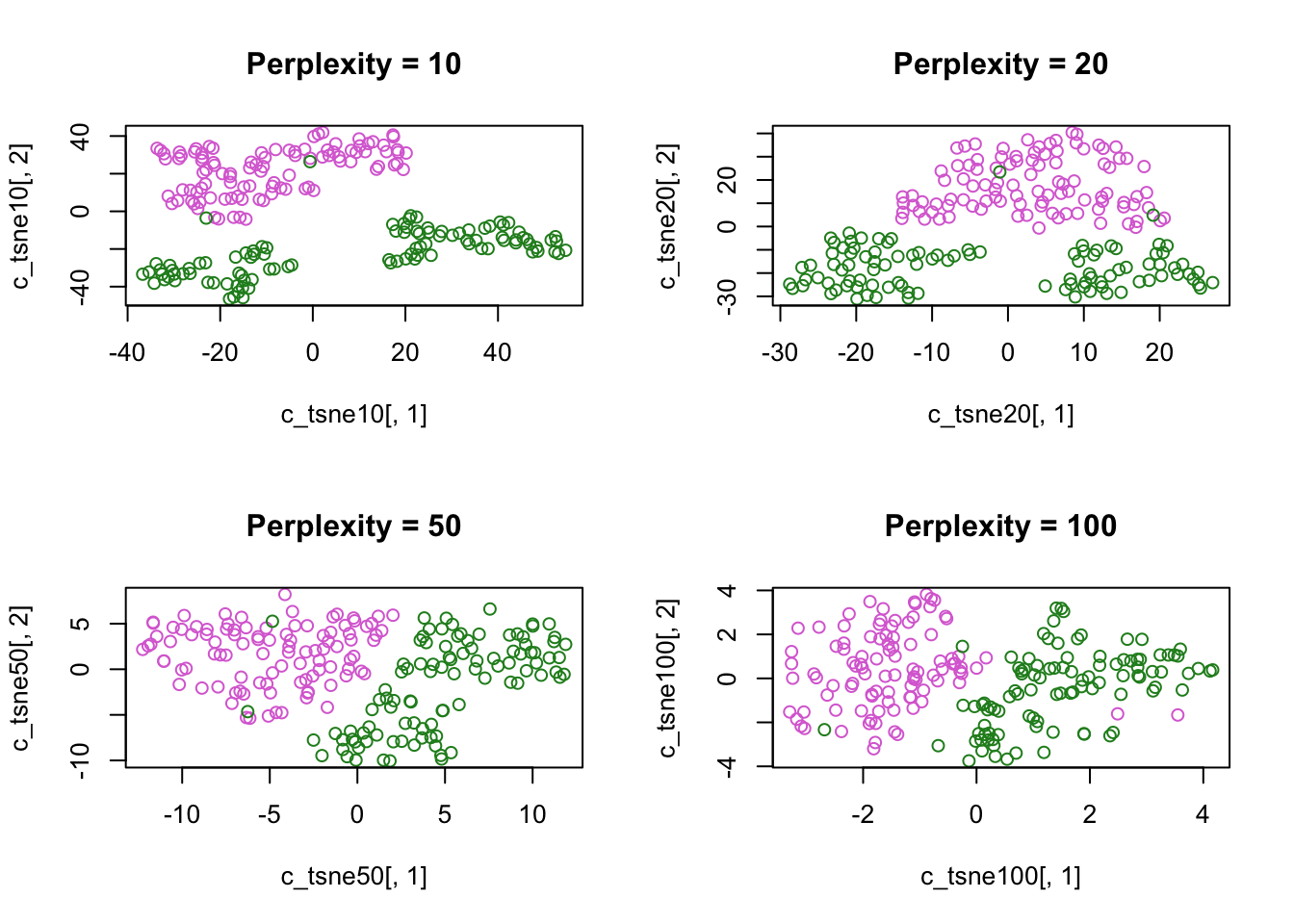
Now do the same, but colour-code for sex:
sex_cols = c("orchid","forestgreen")[factor(crabs$sex)]
par(mfrow=c(2,2))
plot(c_tsne10[,1],
c_tsne10[,2],
main = "Perplexity = 10",
col = sex_cols)
plot(c_tsne20[,1],
c_tsne20[,2],
main = "Perplexity = 20",
col = sex_cols)
plot(c_tsne50[,1],
c_tsne50[,2],
main = "Perplexity = 50",
col = sex_cols)
plot(c_tsne100[,1],
c_tsne100[,2],
main = "Perplexity = 100",
col = sex_cols)
Run UMAP
## List of 4
## $ layout: num [1:200, 1:2] -0.771 -0.723 -0.505 -0.419 -0.219 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:200] "1" "2" "3" "4" ...
## .. ..$ : NULL
## $ data : num [1:200, 1:5] 8.1 8.8 9.2 9.6 9.8 10.8 11.1 11.6 11.8 11.8 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:200] "1" "2" "3" "4" ...
## .. ..$ : chr [1:5] "FL" "RW" "CL" "CW" ...
## $ knn :List of 2
## ..$ indexes : int [1:200, 1:15] 1 2 3 4 5 6 7 8 9 10 ...
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:200] "1" "2" "3" "4" ...
## .. .. ..$ : NULL
## ..$ distances: num [1:200, 1:15] 0 0 0 0 0 0 0 0 0 0 ...
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:200] "1" "2" "3" "4" ...
## .. .. ..$ : NULL
## ..- attr(*, "class")= chr "umap.knn"
## $ config:List of 24
## ..$ n_neighbors : int 15
## ..$ n_components : int 2
## ..$ metric : chr "euclidean"
## ..$ n_epochs : int 200
## ..$ input : chr "data"
## ..$ init : chr "spectral"
## ..$ min_dist : num 0.1
## ..$ set_op_mix_ratio : num 1
## ..$ local_connectivity : num 1
## ..$ bandwidth : num 1
## ..$ alpha : num 1
## ..$ gamma : num 1
## ..$ negative_sample_rate: int 5
## ..$ a : num 1.58
## ..$ b : num 0.895
## ..$ spread : num 1
## ..$ random_state : int 387780946
## ..$ transform_state : int NA
## ..$ knn : logi NA
## ..$ knn_repeats : num 1
## ..$ verbose : logi FALSE
## ..$ umap_learn_args : logi NA
## ..$ method : chr "naive"
## ..$ metric.function :function (m, origin,
## targets)
## ..- attr(*, "class")= chr "umap.config"
## - attr(*, "class")= chr "umap"par(mfrow=c(1,2))
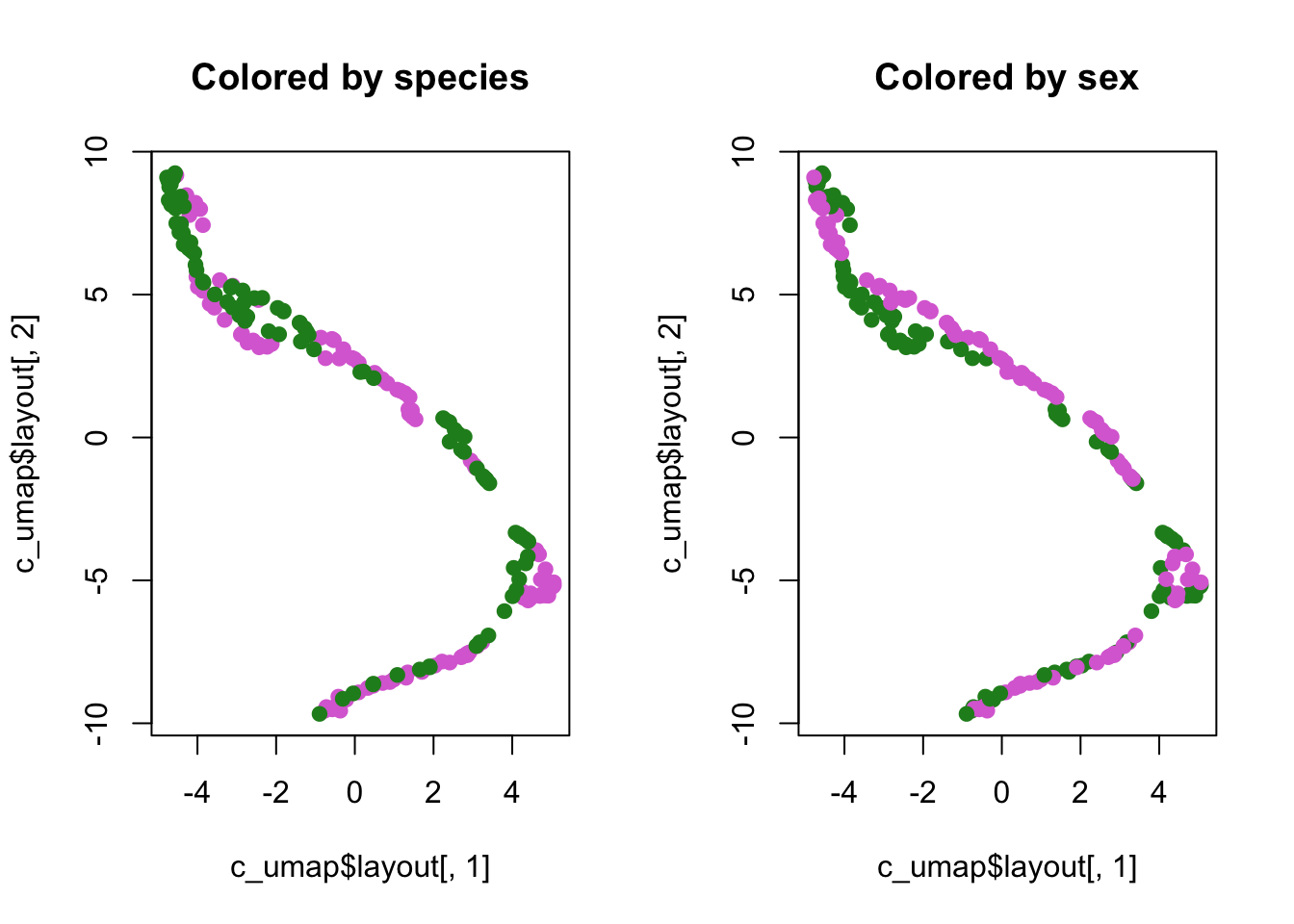
plot(c_umap$layout[,1],
c_umap$layout[,2],
col = species_cols, pch = 19,
main = "Colored by species")
plot(c_umap$layout[,1],
c_umap$layout[,2],
col = sex_cols, pch = 19,
main = "Colored by sex")