Module 4: Exercise Results
if (!requireNamespace("yeastRNASeq", quietly = TRUE))
BiocManager::install("yeastRNASeq")
library(yeastRNASeq)
data(geneLevelData)
gene_keep <- rowMeans(geneLevelData) > 2
# What genes pass this threshold?
filtered <- geneLevelData[gene_keep,]
str(filtered)
## 'data.frame': 6113 obs. of 4 variables:
## $ mut_1: num 38 31 55 29 189 33 23 51 21 4 ...
## $ mut_2: num 39 33 52 26 180 41 13 52 12 5 ...
## $ wt_1 : num 35 40 47 5 151 32 73 54 3 4 ...
## $ wt_2 : num 34 26 47 5 180 29 63 45 3 8 ...
# as.matrix(filtered): the count data in the right class
# phenoData: The sample information
group <- factor(rep(c("Mut", "WT"),each=2), levels = c("WT","Mut"))
y <- DGEList(as.matrix(filtered),
group = group)
## matrix of experimental design
mod = model.matrix(~group, y)
## Normalize data
y <- calcNormFactors(y, method = "upperquartile")
y <- estimateDisp(y, mod)
fit = glmFit(y, mod)
lrt = glmLRT(fit, coef = 2)
diffEx2 <- decideTestsDGE(lrt,
adjust.method="BH",
p.value=0.05
)
table(diffEx2)
## diffEx2
## -1 0 1
## 1282 3147 1684
DEGS = topTags(lrt, n=nrow(y))$table
## check out differentially expressed genes
head(DEGS)
## logFC logCPM LR PValue FDR
## snR59 8.918990 10.146912 2142.162 0 0
## snR49 8.541859 9.773155 1652.824 0 0
## snR41 7.757413 11.217747 4287.880 0 0
## YPL198W 7.499306 10.244671 2224.245 0 0
## snR76 7.191421 9.776288 1602.758 0 0
## snR81 7.028955 10.497704 2595.722 0 0
DEGS_sig = DEGS[DEGS$FDR < 0.05,]
head(DEGS_sig)
## logFC logCPM LR PValue FDR
## snR59 8.918990 10.146912 2142.162 0 0
## snR49 8.541859 9.773155 1652.824 0 0
## snR41 7.757413 11.217747 4287.880 0 0
## YPL198W 7.499306 10.244671 2224.245 0 0
## snR76 7.191421 9.776288 1602.758 0 0
## snR81 7.028955 10.497704 2595.722 0 0
## [1] 2966 5
midx <- match(rownames(DEGS), rownames(diffEx2))
diffEx2 <- diffEx2[midx,]
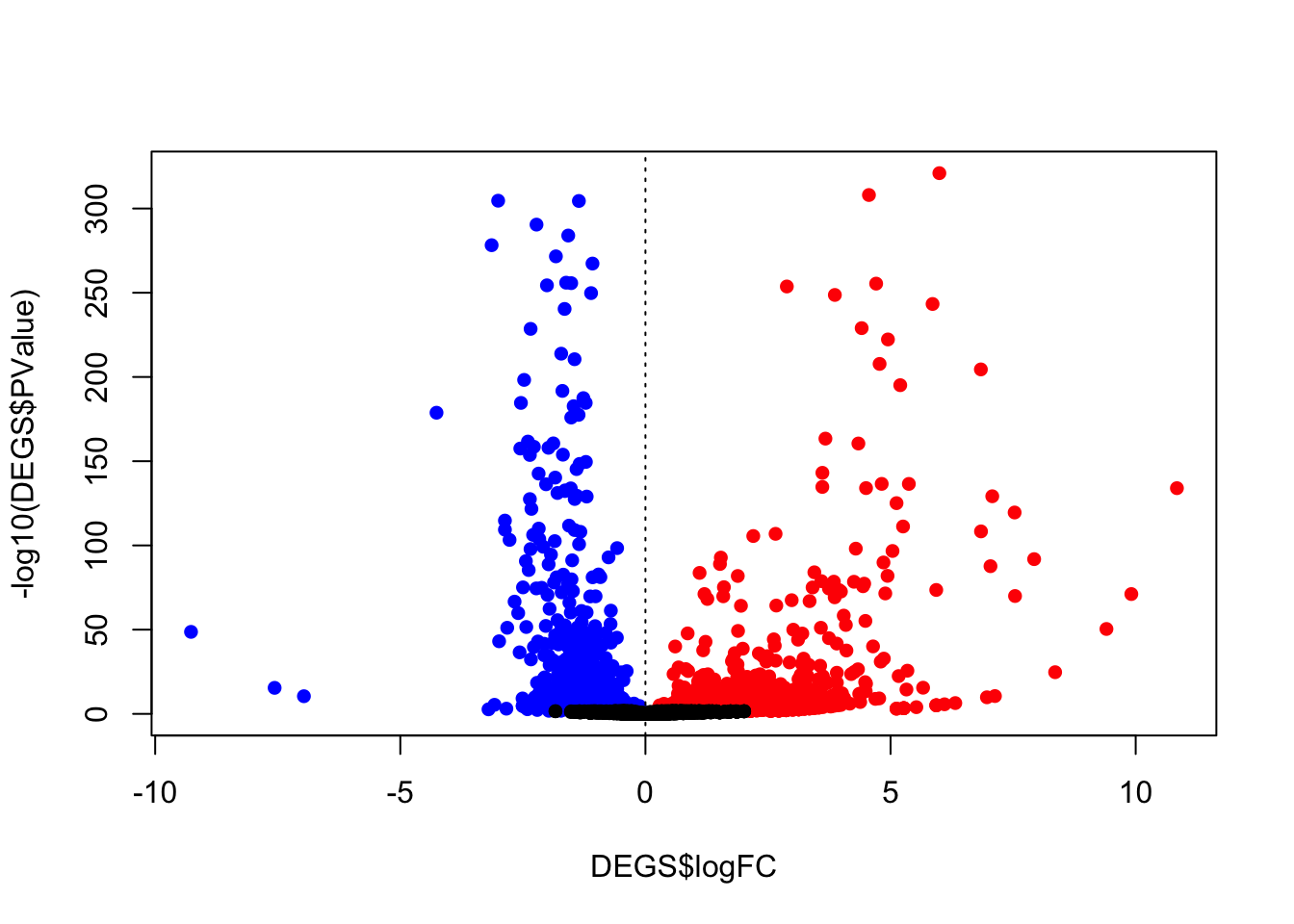
cols <- rep("black",nrow(diffEx2))
cols[which(diffEx2>0 )] <- "red"
cols[which(diffEx2<0)] <- "blue"
# volcano plot
plot(DEGS$logFC,-log10(DEGS$PValue),pch=16,
col=cols)
abline(v=0,lty=3)